A
B
C
D
Omission libraries
Determination of amino acid composition
of bioactive components
of soluble peptide libraries
The number of the component
sub-libraries that have to be synthesized and tested in positional scanning
is quite considerable. In the case of pentapeptides, for example, the number
of sub-libraries in the set is 120. In order to reduce this number we developed
sets of omission libraries that makes possible to determine the sequence
of bioactive peptides present in combinatorial libraries in two steps.
First determine the amino acid composition then determine the sequence
of the previously found amino acids (Furka et al. 1996, Campian et al.
1998).
An omission library
can be prepared by omitting in the synthesis of the library the same amino
acid in all positions. The relation of the omission libraries (B, C and
D) to the full trimer library (A) composed from three amino acids (yellow,
blue and red) in all positions is demonstrated ion the figure.
A
B
C
D
From omission libraries B, C and D the red, blue and yellow amino acids are omitted, respectively. In the component tripeptides of library B, for example, no red amino acid is found. In the synthesis of an alanine omission library alanine is omitted in all positions. As a consequence, no alanine is found in any peptide of the alanine omission library. This makes possible to use the omission libraries in deconvolution. If the full library shows activity and, for example, alanine is present in the bioactive peptide, the activity of the alanine omission library drops relative to that of the full library, since the active peptide is not present.
The applicability of
omission libraries is demonstrated by a model experiment using a tripeptide
amide library for competitive inhibition of binding of LH-RH to its polyclonal
antibody. The tripeptide amide library was synthesized from 20 amino acids.
In construction of the full library, 19 amino acids were used in the first
and second coupling position (cysteine was omitted from all libraries)
and pyroglutamic acid was added to this set for the N-terminal position.
In addition, a full set (19) of omission tripeptide amide libraries, a
tripeptide acid library (omission of the amide group) and a library omitting
pyroglutamic acid was prepared. The libraries were submitted to binding
experiments using radioimmunoassay in determination of binding of radiolabeled
LH-RH to the antibody.
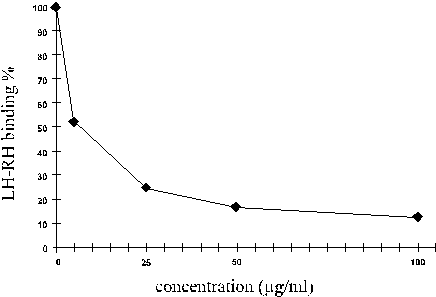
First the full library
was examined. The data of the figure show that the full tripeptide amide
library competes with the radioactively labeled LH-RH in binding to the
antibody since raising the concentration of the library reduces the quantity
of LH-RH bound to the antibody. This result also suggests that the optimal
concentration for further experiments should be around 50 microgram/mL.
Next, the 21 omission libraries were submitted to binding experiments. The figure demonstrates the result. Omission of pyroglutamic acid and the C-terminal amide group is marked by p and -a, respectively. It can be clearly seen that binding of the radio-labeled LH-RH is reduced less by the following omission libraries: -G, -P, and -R.
Effect of omission libraries on binding. X and -a mean full tripeptide
amide and full tripeptide acid libraries,
respectively, while -p denotes pyroglutamic acid omission library.
The other omission libraries are
represented by a minus sign followed by the one letter symbol of the
omitted amino acid. Concentration in
these tests was 50 microgram/mL.
This means that the
amino acids essential for binding are: glycine, proline, and arginine.
The terminal amide group also proved to be essential since the full tripeptide
acid library (-a) does not significantly reduce the LH-RH binding.
The results of the
experiments carried out with omission libraries gave no indication about
the position of the amino acids within the sequence of the active peptide.
Despite this, the information gained by only 21 screening experiments was
very valuable. They defined a library of very low complexity. If an “amino
acid occurrence library” is synthesized by varying only three amino acids,
Gly, Pro, and Arg in all of the three positions, the active tripeptide
is expected to be present among the 27 components of this tripeptide amide
library. In other words, by screening with omission libraries, the complexity
of the library in which the active peptide is found can be reduced from
the original 7220 to only 27.
Positions of the identified
amino acids could be determined by using one of the following three possibilities:
1. Preparation by parallel
synthesis and screening of the 27 components of the occurrence library.
2. Application of
positional scanning to the occurrence library (preparation and screening
of nine sub-libraries).
3. Positional scanning
with nine sub-libraries of the full library (if available).
Since we previously
prepared a full set (58) of sub-libraries for using them in other experiments,
we chose to realize the latter possibility.
In order to determine
the positions of glycine, proline and arginine in the tripeptide responsible
for the competitive binding to the LH-RH antibody, the following sub-libraries
were tested:
1G, 2G, 3G
1P, 2P, 3P
1R, 2R, 3R
G, P, and R are amino acids occupying nonrandomized (1, 2, or 3) coupling positions.
The result of the experiments is shown in the figure.
It can be seen that 3R, 1G, and 2P considerably reduce binding of the labeled
LH-RH to the antibody. This means that glycine, proline and arginine occupy
the first, second and third coupling position in the active tripeptide.
Therefore, the sequence of the tripeptide exhibiting specific binding is
Arg-Pro-Gly-NH2, which happens to be identical with the C-terminal sequence
of LH-RH.
The omission libraries can be easily synthesized and
can be used very effectively in screening of peptide libraries. The amino
acid sequence of the bioactive peptides can be determined, of course, without
omission libraries, for example, by positional scanning. In that case,
however, significantly more screening experiments would have to be performed
(in the above example 58 instead of 29).